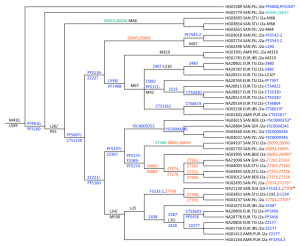
 Under the lead of Greg Magoon a group of citizen scientists (including J2-M172 team members Bonnie Schrack and Chris Rottensteiner) were involved in the analysis of a high-resolution a priori maximum parsimony Y-chromosome (“chrY”) phylogeny created from data of 1292 male samples in the 1000 Genomes Project. 63 of those samples are in haplogroup J2-M172 (42 in J2a and 21 in J2b) and a good picture of the tree structure is available trough the preprint-paper. Chris Rottensteiner was responsible for the J2a part and Vince Tilroe for J2b.
Under the lead of Greg Magoon a group of citizen scientists (including J2-M172 team members Bonnie Schrack and Chris Rottensteiner) were involved in the analysis of a high-resolution a priori maximum parsimony Y-chromosome (“chrY”) phylogeny created from data of 1292 male samples in the 1000 Genomes Project. 63 of those samples are in haplogroup J2-M172 (42 in J2a and 21 in J2b) and a good picture of the tree structure is available trough the preprint-paper. Chris Rottensteiner was responsible for the J2a part and Vince Tilroe for J2b.
SNP markers with names in the range Z6046 through Z8120 were identified and provisionally placed [in J2a …]
J2a-PF4610* is revealed by a Punjabi sample and independently confirmed by seven Sardinian SNP results. By comparisons, this ancient lineage is expected to be defined by Z6046 and its equivalents. […]
J2a-M68 has an Eastern distribution, with two Tamil and one Punjabi samples, and is believed to lie within a haplogroup defined by SNP Z6055 and equivalents, by comparison with two Sardinian samples. This proposed upstream haplogroup J2a-Z6055 could be interesting in future studies of the remaining J2a-L26* haplotype clusters.
J2a-F3133 haplogroup could be probably more reliably described by Z7700 and equivalents. Its subclade, Z7704, has the known subclades L192 and L534 below it.
J2a-PF5174, with three sequences from Tamil, two Bengali and two Punjabi as well as one Gujarati. When 13 Sardinian samples are also considered, a distribution from South Asia to Europe is apparent. Interesting substructure could be revealed by the proposed sub-haplogroups defined by Z6082 and Z7366. Z6082 has the subclade Z7274 further subdivided into Z7255 and Z7261. Z7366, by comparison with three Sardinians, is subdivided in Z6092 and PR2128.
J2a-PF5116,Z2227 sub of PF5087/CTS1230 unites known and new haplogroups with expected expansion mainly to Europe. […] downstream is Z6065 etc. uniting one M47 sample from Punjab and an unknown lineage defined by another two Punjabi samples […]
J2b-M12 now sub-divides into three branches defined by M205, M241, and Z2453.
J2b2b-Z2432 is a significant new clade which exists primarily in south-west Asia, particularly India and surrounding regions.
Generation of high-resolution a priori Y-chromosome phylogenies using “next-generation” sequencing data
An approach for generating high-resolution a priori maximum parsimony Y-chromosome (“chrY”) phylogenies based on SNP and small INDEL variant data from massively-parallel short-read (“next-generation”) sequencing data is described; the tree-generation methodology produces annotations localizing mutations to individual branches of the tree, along with indications of mutation placement uncertainty in cases for which “no-calls” (through lack of mapped reads or otherwise) at particular site precludes a precise placement of the mutation. The approach leverages careful variant site filtering and a novel iterative reweighting procedure to generate high-accuracy trees while considering variants in regions of chrY that had previously been excluded from analyses based on short-read sequencing data. It is argued that the proposed approach is also superior to previous region-based filtering approaches in that it adapts to the quality of the underlying data and will automatically allow the scope of sites considered to expand as the underlying data quality (e.g. through longer read lengths) improves. Key related issues, including calling of genotypes for the hemizygous chrY, reliability of variant results, read mismappings and “heterozygous” genotype calls, and the mutational stability of different variants are discussed and taken into account. The methodology is demonstrated through application to a dataset consisting of 1292 male samples from diverse populations and haplogroups, with the majority coming from low-coverage sequencing by the 1000 Genomes Project. Application of the tree-generation approach to these data produces a tree involving over 120,000 chrY variant sites (about 45,000 sites if “singletons” are excluded). The utility of this approach in refining the Y-chromosome phylogenetic tree is demonstrated by examining results for several haplogroups. The results indicate a number of new branches on the Y-chromosome phylogenetic tree, many of them subdividing known branches, but also including some that inform the presence of additional levels along the “trunk” of the tree. Finally, opportunities for extensions of this phylogenetic analysis approach to other types of genetic data are examined.
