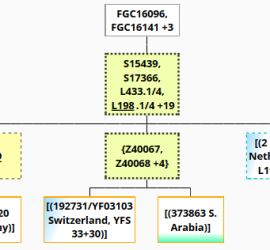
J2-M172 Stats from FTDNA Y-DNA Haplotree (BigY) and YFull YTree
Current subclades (FamilyTreeDNA) and id’s (YFull YTree v11.04.00) analysis. It is biased against customers of Y-clusters or regions of which descendants do more BigY/NGS testing (Ashkenazi/Europe, Northwest Europe, Georgia, Armenia, Rhine/Germans, Arabia, Russia, others Europe, etc.). Most common major subgroups:J2a-L26 73.5%J2a-M67 26.3%J2a-L24 24.8%J2b 20.7%J2a-PF5197 9.1 % Most common (3+%) from the 46 Pre-Chalcolithic J-M172 main sub-haplogroups (defined 2021)J2b-M102>Z529>Z1827>Z593>M241>L283 11.5%J2a-CTS7683>L26>PF5088>PF5160>L24>Z393>L25>Z7700,F3133 11.4%J2a-CTS7683>L26>PF5088>PF5125>Z2227>L558>M67>Z1847>Z7671>CTS3261>CTS6619,Z7661 10.7%J2a-CTS7683>L26>PF5088>PF5160>L24>Z393>L25>Z438 9.2%J2a-CTS7683>L26>PF5088>PF5125>Z2227>L558>M67>Z1847>PF5132>M92 7.1%J2b-M102>Z529>M205 5.3%J2a-CTS7683>L26>PF5088>PF5125>Z2227>Z6065>Y7708>Y7702>M47 4.0%J2a-CTS7683>L26>PF5088>PF5160>L24>Z393>L25>PF4888 3.6%J2a-CTS7683>L26>Z6064>Z6061>Z6057 3.4%J2a-CTS7683>L26>PF5088>PF5160>PF5197>PF5172>Z7314>PF5169>PF5174 3.2%J2a-CTS7683>L26>PF5088>PF5160>PF5197>PF5172>Z7314>PF5169>FGC16096>PH4970,Y14400 3.0% See https://docs.google.com/spreadsheets/d/1Kfvde5gHQ0EVV9lLVnjMLgtyOgLJQPZdywqyu2-idSc/edit?usp=sharing