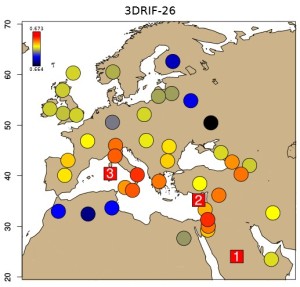
3DRIF-26 IBS to modern populations, 1-3 best matching (SupFig 12 CC-BY Martiniano et al 2016)
The years 2014 and 2015 did provide useful ancient genomes NGS data in an increasing manner (see ancient J2-M172 genomes). For J2 the year 2016 could not begin more exciting: Martiniano et al published a paper about nine ancient genomes from northern Britain: Genomic signals of migration and continuity in Britain before the Anglo-Saxons. As introduction to the sample of interest some quotes from the publication:
Strikingly, one Roman skeleton shows a clear signal of exogenous origin, with affinities pointing towards the Middle East, confirming the cosmopolitan character of the Empire, even at its northernmost fringes. […]
Seven ancient genomes are sampled from a cemetery in Roman York dated between the second and the fourth century AD […]
the majority (6/7) of Driffield Terrace samples belong to sub-lineages of R1b-L52/L11 […] Sample 3DRIF-26, on the other hand, despite belonging to the same burial context, presented a lineage consistent with haplogroup J2-L228 […] majority were adults under 45 years old, male and most had evidence of decapitation. They were slightly taller than average for Roman Britain, displayed a high occurrence of trauma, potentially related to interpersonal violence and evidenced childhood stress and infection. This demographic profile resembles the population structure in a recently excavated burial ground of the second and third century AD at Ephesus, which has been interpreted to be a burial ground for gladiators. However, the evidence could also fit with a military context; the Roman army had a minimum height for recruitment and fallen soldiers would match the young adult profile of the cemetery. […]
Affinity with global populations […] 3DRIF-26 gives a clear Middle Eastern signal, with closest neighbours of Palestinian, Jordanian and Syrian origin. […] ancestral component […] whereas 3DRIF-26 again shows a majority West Asian/Middle Eastern component. Isotopic analyses of the skeletons support this genetic differentiation of 3DRIF-26 from the remainder of the individuals sampled. […] 3DRIF-26, which showed highest IBS with samples from Saudi Arabia. […] assignment of 3DRIF-26 to the Middle East region seems secure, but resolution to an individual population may not be possible. […] imputed lactase persistence genotypes: […] 3DRIF-26 were homozygous for the ancestral non-persistence variant. […]
Discussion: […] affinities clearly lie with the Middle East. Isotopically, the most plausible suggestion is an arid environment on igneous or limestone geology […] Hence, although this individual is indistinguishable from the other inhumations in terms of burial practice and osteology, the analyses show that, even in its northernmost provincial capital, the profoundly cosmopolitan nature of the Roman Empire suggested by documentary and epigraphic sources continued to hold sway […]
Scars from lion bite suggest headless Romans found in York were gladiators is an earlier archaeological article which supports the gladiator theory.
In support of the papers autosomal analysis of 3DRIF-26 the data was analyzed by Davidski/Eurogenes and the Middle Eastern signal is clearly confirmed: Using K15 and oracle 4 populations approximation the nearest modern populations seem to be Samaritans, Tunisian_Jewish, Yemenite_Jewish, Saudis, Egyptians, Cyprians. 1 population approximation lists Palestinian, Bedouin, Jordanian, Samaritan, Egyptian.
A further analysis of 3DRIF-26’s genetic affinities after taking into account his non-trivial Sub-Saharan admixture was done with qpAdm. The best ten models include: Anatolia_Neolithic 0.528, Caucasus_HG Kotias 0.379; Samaritan 0.940, Cypriot 0.915, Lebanese_Druze 0.933, BedouinB 0.998, Lebanese_Christian 0.929, Lebanese_Muslim 0.943, Druze 0.933, Iraqi_Jew 0.924, Roman_outlier 0.900.
these results provide rather convincing evidence that 3DRIF-26’s West Eurasian ancestry is derived from the Levant. Moreover, his relatively high level of Sub-Saharan admixture suggests that he came from the southern Levant or perhaps a nearby region, like the Sinai Peninsula.
Interestingly, the best models feature a couple of religious minorities (Samaritans and Lebanese Druze), an island population (Cypriots), and a fairly unique group in terms of genetic structure from Israel’s Negev Desert (BedouinB). This suggests that 3DRIF-26 may have belonged to a similar religious or geographic isolate population, or, alternatively, that most of the Levant has experienced significant genetic shifts since he was alive.
When asking the corresponding authors Matthew J. Collins and Daniel G. Bradley for help in locating the chrY raw data of 3DRIF-26 shortly after I was kindly provided with the 3DT26.chrY.bam file by Rui Martiniano. Many thanks to them for allowing me to analyze the data so easily.
chrY haplogroup analysis of 3DRIF-26 (3DT26.chrY.bam)
The genome of 3DRIF-26 has a mean coverage ~1.13X which is over the average of the samples. For 3DT26.chrY.bam (9 MB) samtools stats calculates
bases mapped (cigar): 8,585,237
average quality: 38.4
Phylogenetically such coverage should allow to compare Y-SNPs on a low level but still informative for shared paternal lineage ancestry of over 1000 years. The assignment to the J2 haplogroup is confirmed by the derived detection of PF4597. Substream J2a consistently is ancestral (M410, L505) while J2b (M314, Z530) is derived as is J2b1 (M205, CTS560). For certified haplogroups see ISOGG J page.
J2b1-M205 is a rather minority sublineage of J2 and J2b (in J2@FTDNA 72 of 2679 samples, 2.7%) and suffered a long bottleneck before modern diversification supported by YFull age estimates: formed 16200 ybp, TMRCA 5600 ybp (formed CI 95% 18000<->14400 ybp, TMRCA CI 95% 6900<->4300 ybp). The distribution is more similar to J2a sublineages (Middle Eastern with Western and Eastern Expansion) then to those of J2b2 (presence mainly in South Asia and Balkan). Because of the low frequency no informative map is available (YHRD has no derived samples, Singh et al 2016 in the paper about Indian J2 has a J2b1-M205 map which seems to have not enough sampling in the Middle East and especially in Levantic and Arabic areas, see Fig. 2 (b).
For the analysis currently 15 M205+ chrY NGS results are available with varying coverage and data access. Those results so far build 10 subhaplogroups (some tentative) mostly without more then two equivalent SNPs. See tree.j2-m172.info/?Hg=J2b1
3DT26.chrY.bam was ancestral to some of them (PH4306, M280, PF7315/Z1621, YP154, YP56, YP22/Z28761, YP7) many had no-call/s (YP17/Z8764, CTS1969, Y18947, PF7320, PF7370, Z38486, PH1089, PH2734, PH2514, PF7342, YP106).
A non-exhaustive check of novel/private SNPs was also done with no shared derived variant except an ambigous result for Hg19,b37 8197067, G->A, YFS090749, 1G- 1A+ which is derived in N53374/YF01501 Vinnytsia Ukraine 44A+. So unfortunately no more informative matches substream to M205 seem to exist currently to further reconstruct the origin and ancestry of 3DRIF-26 with the following conclusions (see 2020 PF7321 2T+ update at the botton):
- the low-coverage and short chrY length of 3DRIF-26 could fail to detect a relation to some of this basal subhaplogroups, especially the samples derived for Y18947 and the gre-3 Greek Hallast et al 2014 sample also having restricted sequence lenght.
- The lineage of 3DRIF-26 could have died out
- Any sample being M205+ and negative for YP17/Z8764 and CTS1969 (including subgroups) is particularly interesting for further Y-NGS testing, especially if having British paternal lineage ancestry to exclude a relation to 3DRIF-26.
- A NGS resequencing of 3DRIF-26 with higher read depth could facilitate the discovery of shared subhaplogroups of M205
- A standard Y-STR DYS marker sequencing (at least a Y23 panel, preferably the Y37 FTDNA panel) could allow to scan Y-haplotype databases for possibly interesting matches
Analysis Log
J2: G->A, PF4597, 1A+;
J2a: A->G, M410/PF4941, 1A-; G->T, L505/PF4987, 2G-;
J2b: A->C, M314/PF4939, 1C+; T->G, PF7309/Z530, 1G+
J2b2: A->T, Z575, 1A-;
J2b1: T->A, M205, 1A+; G->T, CTS560, 1T+; C->T, PF7308, 1T+;
J2b1 check: 7308226, C->T, CTS1282, 1T+
J2b1 sub ancestral: 19180830, T->A, PH4306, 2T-; 21878762, G->A, M280, 2G-; 23605449, C->T, PF7315/Z1621, 2C-; 16269044, G->A, 1G-; 21812281, A->G, YP154, 1A-; 14784089, A->G, YP56, 2A-; 13958621, G->A, YP22/Z28761, 1G-; 6729677, G->T, YP7, 1G-;
J2b1 sub ambiguous: N53374 Novel, 8197067, G->A, YFS090749, 1G- 1A+;
no call/s: YP17/Z8764, CTS1969, Y18947, PF7320, PF7370, Z38486, PH1089, PH2734, PH2514, PF7342, YP106;
Discoveries and statements by Ted Kandell (OGF) about 3DRIF-26:
an extinct mtDNA H5* C5349T C6041T and H5* isn’t found in Arabia or Egypt.
[Davidski/Eurogenes qpAdm:] His Anatolian Neolithic is 52.8%, extremely high, the same as Sardinians who are 50%-60%. His next best fit, pretty close, is Samaritan 94% Yoruba 6%. After that the chi squared gets much higher and the probability rapidly decreases.
Also, he wasn’t beheaded but his head was bashed in the “coup de grace” given to defeated gladiators. He wasn’t buried with grave goods, either.
“J2e” M314/M12 xM241 was found by Shen et al. (2004) in 2 of 20 (10%) of Yemenite Jews
Jan. 2020 analysis by Flor Veseli
He is negative on SNPs that were covered for every M205>PF7321 subclade, including negative on the Private SNPs of the Ukrainian J-PF7321*, except for CTS1969 which has no reads. So he at J-PF7321* or J-CTS1969* in case CTS1969+. Below are some important SNP calls:
PF7321 2T+
Y3163 1G+
Y101509 2A-
YP51 1C-
CTS1969 ?? (no reads)
Y45447 2T-
Y27394 2G-
BY88216 2G-
CTS10179 2T-
Y22521 1T-
FT45285 1G-
Y134194 1G-
Y134202 1G-
FT45279 2T-