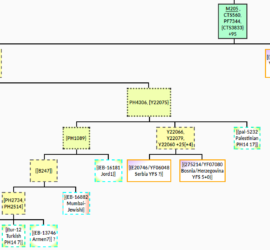
possibly J2b1-M205 basal structure defined: CTS1969,CTS5338,YP13,PH4306,PH1089,Y22066
Further analysis of NextGenSequences at YFull (see yfull.com/tree/J-M205/) combined with results from scientific studies (SNP, VCF & BAM results) has revealed a more consistent and hopefully informative basal substructure for J-M205 (see tree.j2-m172.info/?Hg=J2b1) then what was available at the latest major update: J2b-M205>PH4306: a little substructure for this isolated lineage. See also yfull.com/tree/J-M205/ It seems likely that the many remaining […]