(UPDATE 2015-05-04: three FGC Wael Y-sequences) Since the first Geno 2.0 results were known for the old J2a4i (L198,L433) and the yet uncertified L1064 clusters, we did know they were PF5169(xPF5174) which means positive for PF5169 and negative for PF5174. Since not many J2 subclades have more then two parallel “brother” subclades I suspected that J2a-L198,L433 and L1064 shared some SNPs. When I first analyzed Y-data from Genome of the Netherlands data (GoNL) project in July 2014 I found 4 samples positive for PF5169 but only two for PF5174. Additionally two samples were positive for L198. At the same time we finally got the first high-res Y-sequence from a PF5169(xPF5174) kit: a FGC Y-Elite of FTDNA kit N37321 Taylor. Comparing to the GoNL I found 6 SNPs in the private set of FGC-P9ZTZ Taylor (FGC16008-FGC16161) that are shared with two dutch GoNL men:
FGC16064, FGC16081, FGC16096, FGC16105.1/2?, FGC16141, FGC16152. FGC16105 was excluded as recurrent or unreliable, but the other five seemed good.
Fortunately in February/March 2015 we got four additional Y-sequences (one FGC Y-Elite, three FTDNA BigY) including one J2a-L198,L433 result (192731 Schrack, son of 9856). That allowed to certify the following SNPs as shared with L1064: FGC16096, FGC16141, FGC16152. FGC16064/Y14370 is still under investigation as maybe in a bad region or uncovered by BigY currently. FGC16081 is classified as unreliable.
See FGC16096 Research Tree simplified.
Ancient relation analysis: FGC16096, L198,L433, L1064
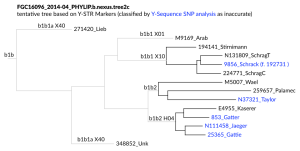
What is most interesting from the analysis of the results is the phylogeny, meaning the reconstruction of the paternal pedigree based on Y-SNPs which for splits based on 2 or more SNPs is very confident and solid. The comparison with Y-STR marker distance predictions clearly shows the great range of error using the up- and down mutating values.
When using YFull YTree age estimates FGC16096 is ca. 11,665 years old and the split between P9ZTZ/N37321 Taylor (incl. the Wael kits) and NUXNR/25365 Gattie 5,300 years ago. That is substantially more then Y-STR distance estimations calculate. Using a calibrated mutation-rate excluding 9 fast mutating markers I had to set the Y-Utility Probability to 99% and the years/generation to 36 to have a distance of the Gattie/Gatter cluster to N37321 Taylor of approx. 4,600 years. But the distance to Schrack/Schrag can’t be also approximated good enough: it was only ca. 7,450 years and the distance to other clusters in L198,L433 is more around 5,400 years. So it seems from Y-STRs with the used methodology it is near to impossible to predict anything very useful regarding the phylogeny over a distance of more then 3,000 years.
Cluster relation analysis: L1064 Gattie/Gatter
If we look at the clear Gattie/Gatter cluster with the used “ancient” Y-STR presetting we get distances from 1,100 to 2,100 years. The latter seems an overestimation.
Genetic distances: 25365 Gattie vs 853 Gatter: GD 6/37 (16%), 25365 Gattie vs N111458 Jäger: GD 4/37 (11%), N111458 Jäger vs 853 Gatter: GD 7/37 (19%),
If we compare to the Y-SNP phylogeny there is one surprise: while by Y-STR28 (fast mutating markers excluded) N111458 Jaeger seems nearer to 25365 Gattie (1,110 years) the Y-SNP analysis clearly shows N111458 Jäger shares 6 SNPs with 853 Gatter which are negative in 25365 Gattie. So we have a inverted phylogeny for this three kits. N111458 Jäger has genealogy origins 1729 in Leimen, Südwestpfalz (SW Palatinate) and from Peer Gatter’s research the 853 Gatter (New York/Boston) lineage could go back very well to one of the historical Gatter regions in Palatinate or as well Alsace. The genealogy origin of 25365 Gattie is 1700 Seltz, Alsace. So all of the cluster seems to have its origins in the Upper Rhine area. Interesting would be to find the reason why Gattie/Gatter share a similar surname, while a later descendant changed the surname to Jäger (hunter).
The SNP age estimates from my manual analysis have a greater confidence then those based on STR, at least the phylogeny (pedigree) relations are solid. I would however like to get YFull analysis of 853 Gatter and N111458 Jäger for a better confidence of terminal SNPs (shared and personal)
Current certifiable phylogeny (ISOGG proposal)
• • • • • • • • • • J2a1a1a2b1b FGC16096, FGC16141, FGC16152
• • • • • • • • • • • J2a1a1a2b1b* –
• • • • • • • • • • • J2a1a1a2b1b1 L433.1/M4853.1, L88.2/L431.1, L198.1
• • • • • • • • • • • • J2a1a1a2b1b1* –
• • • • • • • • • • • • J2a1a1a2b1b1a M419
• • • • • • • • • • • J2a1a1a2b1b2 L1064, FGC16008/Y14562, FGC16009/Y14351, FGC16013/Y14564, FGC16014/Y14565, FGC16034/Y14358, FGC16046/Y14363, FGC16049/Y14570, FGC16052/Y14364, FGC16053/Y14571, FGC16054/Y14572, FGC16056/Y14366, FGC16057/Y14367, FGC16058/Y14368, FGC16059/Y14573, FGC16062/Y14369, FGC16066, FGC16069, FGC16071/Y14371, FGC16073/Y14372, FGC16080/Y14375, FGC16084/Y14376, FGC16085/Y14576, FGC16095, FGC16097/Y14378, FGC16099/Y14379, FGC16100/Y14577, FGC16102/Y14381, FGC16112/Y14387, FGC16115/Y14388, FGC16116/Y14389, FGC16121/Y14390, FGC16123/Y14391, FGC16124/Y14392, FGC16125/Y14563, FGC16128/Y14394, FGC16129/Y14395, FGC16131/Y14578, FGC16133/Y14579, FGC16134/Y14396, FGC16137/Y14397, FGC16140/Y14581, FGC16142/Y14582, FGC16143/Y14400, FGC16146/Y14401, FGC16147/Y14584, FGC16148/Y14402, FGC16155/Y14585, FGC16158/Y14403, FGC30757/Y14365, FGC30774/Y14377, FGC30801, L250.2, PH4968/Y14398/Z32180, PH4970/Y14399/Z32181, Y14353, Y14569/Z34433
• • • • • • • • • • • • J2a1a1a2b1b2* –
• • • • • • • • • • • • J2a1a1a2b1b2a FGC16088, FGC16079, FGC16130, FGC16157
• • • • • • • • • • • • • J2a1a1a2b1b2a* –
• • • • • • • • • • • • • J2a1a1a2b1b2a1 FGC35000, FGC34962, FGC34963, FGC34964, FGC34965, FGC34969, FGC34970, FGC34971, FGC34972, FGC34973, FGC34974, FGC34976, FGC34977, FGC34978, FGC34979, FGC34980, FGC34981, FGC34982, FGC34984, FGC34986, FGC34987, FGC34988, FGC34990, FGC34991, FGC34992, FGC34993, FGC34994, FGC34995, FGC34996, FGC34997, FGC34998, FGC34999, FGC35001, FGC35002, FGC35003, FGC35004, FGC35005, FGC35006, FGC35008, FGC35009, FGC35011, FGC35012, FGC35013, FGC35014
• • • • • • • • • • • • J2a1a1a2b1b2b FGC30775, FGC30743, FGC30753, FGC30754, FGC30755, FGC30760, FGC30761, FGC30762, FGC30764, FGC30765, FGC30767, FGC30771, FGC30776, FGC30778, FGC30780, FGC30782, FGC30784, FGC30785, FGC30786, FGC30788, FGC30789, FGC30792, FGC30793, FGC30797, FGC30799, FGC30803, FGC30804, Z34434, Z34435, Z34436, Z34437
• • • • • • • • • • • • • J2a1a1a2b1b2b* –
• • • • • • • • • • • • • J2a1a1a2b1b2b1 Z34440, Z34438, Z34439, Z34442, Z34443, Z34444
Most useful and interesting To Do’s
- Include all available Y-sequence results in the YFull analysis and YTree
- Testing of one or more so far unique SNPs shared by 192731 Schrack and 2 GoNL in a M419+ sample: S15439, S17366, S19509, S23510, Z32170, Z32171, Z32172, Z32173, Z32174, Z32175, Z32176, Z32178, Z32179
- Testing of one member of the L433 branch with a Y-sequence test of FGC Y-Elite or better resolution (estimated to reveal at least 41 more informative SNPs for certification by comparison with the L1064 branch)
- Testing of as much members of different L433 and L1064 clusters as possible with BigY or better FGC Y-Elite resolution.
- Validating 1-2 SNPs per haplogroup by sanger sequencing (YSEQ) and use them as primary haplogroup naming SNPs.
Analysis Files
Accessible under drive.google.com/folderview…