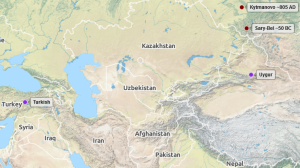
 The huge Allentoft et al. 2015 research with low-coverage genomes from 101 ancient humans from across Eurasia included two males from Altai Iron Age (900 BC to 1000 AD) in the same J2a2 haplogroup (see Altai archaeology). Even more intriguing is that the ancient genomes share unique Y-SNPs with two modern samples: one Uygur and one Turkish, currently best named as J2a2-PH3085,SK1403. Unfortunately only three J2a2 Y-sequences with good coverage are known, two of them at processed at YFull: J2a2-PF5050(xPF5058) HG01589 Punjabi and J2a2-M340 YF01485 Armenian. The third is likely a J2a2-Z35890,P279(xM340,Z28408) from Kuwait. So currently only the diversification of J2a2-PF5050 has a good estimate: 16200 ybp. As long as we have no good age estimates of J2a2-PF5056,PF4993 there will remain much speculation about the age, origin and migration of J2a2-PH3085,SK1403. Could be of classic J2a Fertile Crescent area origin including a migration to Altai/Uygur region with “back-migration” to modern Turkey or the opposite: an origin in Central Asia. Without better data I would however tend to assume the origin of J2a2-PH3085,SK1403 in the South or Eastern Caspian area with migrations to the East and the West.
The huge Allentoft et al. 2015 research with low-coverage genomes from 101 ancient humans from across Eurasia included two males from Altai Iron Age (900 BC to 1000 AD) in the same J2a2 haplogroup (see Altai archaeology). Even more intriguing is that the ancient genomes share unique Y-SNPs with two modern samples: one Uygur and one Turkish, currently best named as J2a2-PH3085,SK1403. Unfortunately only three J2a2 Y-sequences with good coverage are known, two of them at processed at YFull: J2a2-PF5050(xPF5058) HG01589 Punjabi and J2a2-M340 YF01485 Armenian. The third is likely a J2a2-Z35890,P279(xM340,Z28408) from Kuwait. So currently only the diversification of J2a2-PF5050 has a good estimate: 16200 ybp. As long as we have no good age estimates of J2a2-PF5056,PF4993 there will remain much speculation about the age, origin and migration of J2a2-PH3085,SK1403. Could be of classic J2a Fertile Crescent area origin including a migration to Altai/Uygur region with “back-migration” to modern Turkey or the opposite: an origin in Central Asia. Without better data I would however tend to assume the origin of J2a2-PH3085,SK1403 in the South or Eastern Caspian area with migrations to the East and the West.
Phylogeny Analysis (SNPs, Haplogroups)
Sary-Bel Kurgan burial sample (RISE602, SAMEA3325434, ERX925332). From archaeological descriptions it looks like complexes (II c. BC – I century AD) Bulan-Koba culture, which has been following the Scythian-Saka epoch in the Russian Altai (near border to Kazakhstan, China, Mongolia).
RISE602 is positive for (Aburto BAM Analysis Kit file): M410+ CTS7683- PF5058+ PF5073+ PF5000+ PF5032+ PF5037+ PF5017+ PF5027+ PF5052+ PF5060+ PF7380+ PF5027+ P279- M340- PF5048- PF5061-
Kytmanovo 721-889 AD (RISE504, SAMEA3325397, ERX925295) also from the Russian Altai (more northern then RISE602 / south of Novosibirsk).
RISE504 is positive for the following J2a2 and below SNPs (more comprehensive BAM analysis): PF5022+ PF5050+ Z35775+ PF5035+ PF5041+ PF5073+ L581+ PF5044+ PF5000+ PF5002+ PF5013+ PF5031+ PF5032+ PF5038+ PF7384+ PF5015+ PF5017+ PF5027+ PF5036+ PF5043+ PF5056+ PF5064+ PF5003+ PF5027+ PF5031+ Z35889+ Z28380+ Z28381+ Z28386+ Z28401+ Z28402+ Z28403+ Z28420- Z28424- P279.1- YFS082471- Z35890- YFS082495+ YFS082526- YFS082653+ YFS083128- YFS083130+ M340- PF5061- PF5067- PF5070- PF5071- PF5082- L383.1-
 Since PF5056 is equivalent to PF5060 and PF5048 is equivalent to PF5082 they are in the same paragroup.
Since PF5056 is equivalent to PF5060 and PF5048 is equivalent to PF5082 they are in the same paragroup.
Analysis of “singleton” SNPs was done only by comparison with two other PF5056(xP279,PF5048) samples because of the low coverage of the ancient samples (risk of false detection). The samples have different coverage:
- RISE602 Sary-Bel, BAM,
PH1105+ PH1678+ PH1984+ PH2125+ PH3085+ PH3766+ PH3934+ PH5426+ SK1403+ - RISE504 Kytmanov, BAM,
PH358+ PH562+ PH638+ PH1984+ PH2125+ PH3085+ PH4480+ SK1403+ Z19832+ - Uyg1304 Uygur, SK-SNPs/Stoneking BAM, 0.5 Mb, Lippold et al 2014, HGDP01304
PH2466+ SK1403+ SK1404+ SK1405+
- Tur-13 Turkey, PH-SNPs list, 3.5 Mb, Hallast et al 2014, Y-STR23 available
All PH-SNPs listed above positive and additionally SK1403+ SK1404+ SK1405+ Z19832+ Four additional PH-SNPs are negative at least in one of the above samples and 10 additional PH-SNPs with no-reads in the other three samples
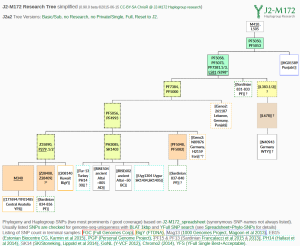
Shared Y-Haplogroup: J2a2-PF5050>PF5058>PF5000>PF5056>PH3085,SK1403 or shorter form J2a2-PH3085,SK1403
See http://tree.j2-m172.info/?Hg=J2a2
The “L24 group” did collect RISE602 info (Людмила Рябченко translation, Kemal AL Gazzah with Ted Kandell first BAM analysis, Al Aburto usage of BAM Analysis Kit on the BAM file (SNP extraction) and David Dugas the first prediction of the J2a2 position.
STR-Haplotype Analysis
Based on a comparative analysis using the Tur-13 Turkey haplotype the following J2@FTDNA group was created:
J2a PF5050>PF5058>PF7384>PF5056>PH3085,SK1403>PH-tur-13 & GD≤5/18
Genetic Distance Table of the haplotypes in this txt file (FTDNA format)
| 1 | modal (Diagonal line is the number of markers) | 49 | 2 | 2 | 2 | 4 | 2 | 4 | 5 | 4 | 2 | 3 | 5 | 6 | 9 | 7 | 7 | 7 |
| 2 | tur-13 Turkish Hallast et al. 2014, this study J2a2-PH3085,SK1403 | 2 | 23 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 4 | 5 | 4 | 5 |
| 3 | TK9_69 Turkmen-Jawzjan Di Cristofaro et al. 2013 J2a-M410(xL26) | 2 | 1 | 39 | 2 | 4 | 2 | 6 | 5 | 6 | 4 | 3 | 6 | 5 | 3 | 4 | 6 | 6 |
| 4 | TK9_7 Turkmen-Jawzjan Di Cristofaro et al. 2013 J2a-M410(xL26) | 2 | 1 | 2 | 39 | 6 | 4 | 4 | 5 | 6 | 4 | 3 | 4 | 3 | 4 | 5 | 6 | 4 |
| 5 | TJ1_36 Tajik-Balkh Di Cristofaro et al. 2013 J2a-M410(xL26) | 4 | 1 | 4 | 6 | 39 | 3 | 8 | 7 | 8 | 6 | 5 | 6 | 6 | 3 | 6 | 5 | 6 |
| 6 | UZ1_4 Uzbek-Balkh Di Cristofaro et al. 2013 J2a-M410(xL26) | 2 | 2 | 2 | 4 | 3 | 34 | 5 | 5 | 1 | 4 | 2 | 6 | 5 | 3 | 5 | 4 | 5 |
| 7 | TJ5_13 Tajik-Badakhshan Di Cristofaro et al. 2013 J2a-M410(xL26) | 4 | 2 | 6 | 4 | 8 | 5 | 39 | 8 | 7 | 4 | 6 | 7 | 5 | 6 | 7 | 5 | 4 |
| 8 | TJ5_6 Tajik-Badakhshan Di Cristofaro et al. 2013 J2a-M410(xL26) | 5 | 2 | 5 | 5 | 7 | 5 | 8 | 39 | 8 | 7 | 3 | 7 | 6 | 4 | 7 | 4 | 6 |
| 9 | IR1_12 East Azeri Di Cristofaro et al. 2013 J2a-M410(xL26) | 4 | 2 | 6 | 6 | 8 | 1 | 7 | 8 | 42 | 6 | 5 | 9 | 7 | 7 | 7 | 7 | 5 |
| 10 | MG3_55 Mongol-NorthWest Di Cristofaro et al. 2013 J2a-M410(xL26) | 2 | 2 | 4 | 4 | 6 | 4 | 4 | 7 | 6 | 43 | 5 | 7 | 6 | 6 | 6 | 7 | 5 |
| 11 | MG3_54 Mongol-NorthWest Di Cristofaro et al. 2013 J2a-M410(xL26) | 3 | 2 | 3 | 3 | 5 | 2 | 6 | 3 | 5 | 5 | 43 | 5 | 6 | 5 | 7 | 6 | 6 |
| 12 | U2 Uyghur Di Cristofaro et al. 2013 J2a-M410(xL26) | 5 | 2 | 6 | 4 | 6 | 6 | 7 | 7 | 9 | 7 | 5 | 43 | 6 | 7 | 9 | 9 | 6 |
| 13 | 321244 Khasauov, Balkaria (Malkar, Mukhol) Unknown Origin J2-M172 | 6 | 3 | 5 | 3 | 6 | 5 | 5 | 6 | 7 | 6 | 6 | 6 | 37 | 12 | 11 | 11 | 8 |
| 14 | 249616 nayman, koban-noghay, NGY Russian Federation J-M172 | 9 | 4 | 3 | 4 | 3 | 3 | 6 | 4 | 7 | 6 | 5 | 7 | 12 | 37 | 8 | 11 | 11 |
| 15 | 250596 Kazakhstan J-M172 | 7 | 5 | 4 | 5 | 6 | 5 | 7 | 7 | 7 | 6 | 7 | 9 | 11 | 8 | 37 | 11 | 11 |
| 16 | N40835 Russian Federation J-M410 | 7 | 4 | 6 | 6 | 5 | 4 | 5 | 4 | 7 | 7 | 6 | 9 | 11 | 11 | 11 | 67 | 14 |
| 17 | 119738 Mityahdin,b.c.18..,Tatarskiye Junki Russian Federation J-M410 | 7 | 5 | 6 | 4 | 6 | 5 | 4 | 6 | 5 | 5 | 6 | 6 | 8 | 11 | 11 | 14 | 67 |
