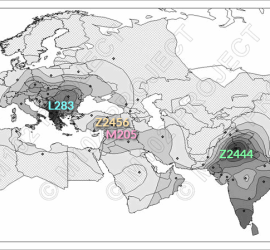
Bekada et al 2015 have published their paper “Genetic Heterogeneity in Algerian Human Populations”. In the present Algerian study only one J2a -M410 was found in North-west Oran1. Combined with earlier studies 8 J2 samples out of 305 total (Y-STR19 haplotypes + major Y-SNPs, S1 Table). By comparison there are 54 J1-M267 samples. IMHO another puzzle piece making early J2 expansion over Northwest Africa to Iberia/Europe unlikely. Other routes seem to be better expansion vectors.
More terminal position of former J2a-L26* Cluster F*
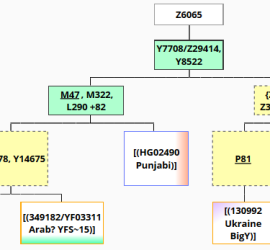
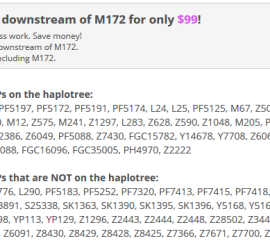
Position found trough a Y-STR67 match not in the J2 project being J-Y7708: J2a-L26>PF5087>PF5116>Z2227>Z6065>Y7708>Z39478?(xM47,P81) One of the last bigger haplotype clusters existing at the begin of 2013 (before Geno 2.0) has found an informative place. See one of my earliest studies figshare.com/articles/J2a_M410_subclades_and_clusters_overview/106387 Thanks to the early J2 haplotype cluster researchers Bonnie Schrack, Angela Cone, wo did the founding grouping. Surnames […]