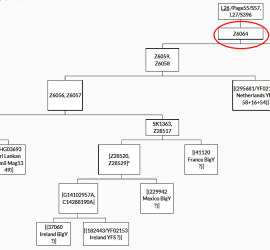
Substantial new subgroups for Z467: S25258 and Z447,Z458(xL210)
As posted in the previous posts substream of the J2a-M67 haplogroup other new findings were possible lately. The most substantial refinement of the M67 tree was possible in the Z467 subclade using 8 BigY results, one of them (B1300/YF02101 Ireland) with additional YFull analysis, gre-77 Greek Hallast et al 2014, Sardinian #845 Francalacci et al 2015, 4 Genome of the […]